-Search query
-Search result
Showing 1 - 50 of 90 items for (author: newman & a)

EMDB-16328: 
Outer membrane attachment porin OmpM1 from Veillonella parvula
Method: single particle / : Silale A, van den Berg B

EMDB-16332: 
Outer membrane attachment porin OmpM1 from Veillonella parvula, native
Method: single particle / : Silale A, van den Berg B

EMDB-16333: 
Outer membrane attachment porin OmpM1 from Veillonella parvula, C3 symmetry
Method: single particle / : Silale A, van den Berg B

PDB-8bym: 
Outer membrane attachment porin OmpM1 from Veillonella parvula
Method: single particle / : Silale A, van den Berg B

PDB-8bys: 
Outer membrane attachment porin OmpM1 from Veillonella parvula, native
Method: single particle / : Silale A, van den Berg B

PDB-8byt: 
Outer membrane attachment porin OmpM1 from Veillonella parvula, C3 symmetry
Method: single particle / : Silale A, van den Berg B

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-8dtk: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P
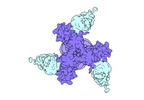
EMDB-27706: 
Vaccine elicited Antibody MU89 bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer
Method: single particle / : Stalls V, Acharya P
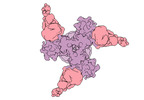
EMDB-27776: 
Vaccine elicited Antibody MU89+S27Y bound to CH848.D949.10.17_N133D_N138T.DS.SOSIP.664 HIV-1 Env trimer
Method: single particle / : Stalls V, Acharya P

EMDB-15971: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

PDB-8bcz: 
SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45
Method: single particle / : Duyvesteyn HME, Ren J, Stuart DI

EMDB-27267: 
CryoEM structures of bAE1 captured in multiple states.
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I

EMDB-27856: 
CryoEM structures of bAE1 captured in multiple states
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I
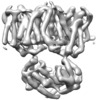
EMDB-28055: 
CryoEM structures of bAE1 captured in multiple states.
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I

PDB-8d9n: 
CryoEM structures of bAE1 captured in multiple states.
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I

PDB-8e34: 
CryoEM structures of bAE1 captured in multiple states
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I

PDB-8eeq: 
CryoEM structures of bAE1 captured in multiple states.
Method: single particle / : Zhekova HR, Wang WG, Jiang JS, Tsirulnikov K, Muhammad-Khan GH, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Tieleman P, Zhou ZH, Pushkin A, Kurtz I

EMDB-24444: 
Mouse GITR (mGITR) with DTA-1 Fab fragment
Method: single particle / : Meyerson JR, He C

PDB-7rfp: 
Mouse GITR (mGITR) with DTA-1 Fab fragment
Method: single particle / : Meyerson JR, He C

EMDB-24683: 
Cryo-EM Structure of the Sodium-driven Chloride/Bicarbonate Exchanger NDCBE (SLC4A8)
Method: single particle / : Wang WG, Tsirulnikov K, Zhekova H, Kayik G, Muhammad-Khan H, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Zhou ZH, Pushkin A, Kurtz I

PDB-7rtm: 
Cryo-EM Structure of the Sodium-driven Chloride/Bicarbonate Exchanger NDCBE (SLC4A8)
Method: single particle / : Wang WG, Tsirulnikov K, Zhekova H, Kayik G, Muhammad-Khan H, Azimov R, Abuladze N, Kao L, Newman D, Noskov SY, Zhou ZH, Pushkin A, Kurtz I

EMDB-23545: 
Cryo-EM structure of the wild-type human serotonin transporter complexed with vilazodone, imipramine and 15B8 Fab
Method: single particle / : Yang D, Kalenderoglou IE, Gouaux E, Coleman JA, Loland CJ

PDB-7lwd: 
Cryo-EM structure of the wild-type human serotonin transporter complexed with vilazodone, imipramine and 15B8 Fab
Method: single particle / : Yang D, Kalenderoglou IE, Gouaux E, Coleman JA, Loland CJ

EMDB-11997: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

PDB-7b3y: 
Structure of a nanoparticle for a COVID-19 vaccine candidate
Method: single particle / : Duyvesteyn HME, Stuart DI

EMDB-10182: 
Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc with cardiolipin
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

EMDB-10183: 
Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

EMDB-10184: 
Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc with cardiolipin
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

EMDB-10185: 
Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

PDB-6sgr: 
Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc with cardiolipin
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

PDB-6sgs: 
Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

PDB-6sgt: 
Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc with cardiolipin
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

PDB-6sgu: 
Cryo-EM structure of Escherichia coli AcrB and DARPin in Saposin A-nanodisc
Method: single particle / : Szewczak-Harris A, Du D, Newman C, Neuberger A, Luisi BF

EMDB-4730: 
Subtomogram average of MAC sheath and inner tube of P. luteoviolacea Mif1 mutant
Method: subtomogram averaging / : Eisenstein F, Medeiros J, Pilhofer M

EMDB-4731: 
Subtomogram average of MAC sheath and inner tube of wildtype P. luteoviolacea
Method: subtomogram averaging / : Eisenstein F, Medeiros J, Pilhofer M

EMDB-20074: 
Chimpanzee SIV Env trimeric ectodomain.
Method: single particle / : Pallesen J, Andrabi R, de Val N, Burton DR, Ward AB

PDB-6ohy: 
Chimpanzee SIV Env trimeric ectodomain.
Method: single particle / : Pallesen J, Andrabi R, de Val N, Burton DR, Ward AB

EMDB-4526: 
Human post-catalytic spliceosome (P complex) stalled with DHX8 S717A mutant, overall map
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4527: 
Human post-catalytic spliceosome (P complex) stalled with DHX8 K594A mutant, focused refinement on core
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K

EMDB-4528: 
Human post-catalytic spliceosome (P complex) stalled with DHX8 S717A mutant, focused refinement on core
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4529: 
Human post-catalytic spliceosome (P complex), focused refinement on Aquarius and SYF1
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4530: 
Human post-catalytic spliceosome (P complex), focused refinement on Brr2
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4532: 
Human post-catalytic spliceosome (P complex), focused refinement on DHX8
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4533: 
Human post-catalytic spliceosome (P complex), focused refinement on Prp19
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4534: 
Human post-catalytic spliceosome (P complex), focused refinement on U2 snRNP
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4535: 
Human post-catalytic spliceosome (P complex), focused refinement on U5 Sm
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai KN

EMDB-4525: 
Human post-catalytic spliceosome (P complex) stalled with DHX8 K594A mutant, overall map
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K

PDB-6qdv: 
Human post-catalytic P complex spliceosome
Method: single particle / : Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K

EMDB-0173: 
Complex of foot-and-mouth-disease virus (type O1 M) with the Fab of antibody D9. The virus is at 3.5 Ang resolution, the Fab is flexibly attached and at much lower resolution.
Method: single particle / : Shimmon G, Kotecha A, Ren J, Asfor AS, Newman J, Berryman S, Cottam EM, Gold S, Tuthill TJ, King DP, Brocchi E, King AMQ, Owens R, Fry EE, Stuart DI, Burman A, Jackson T
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
